| HOME | METHODS | LINKS | NEWS | POLLS | FORUM | CONTACT | SEARCH |
| PCR for PDH | ||||||||||||||||||||||||||||||||||
|
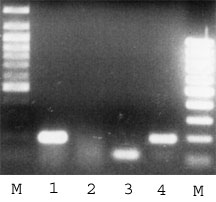
Amplification of 102 bp fragment from PDH cDNA and about 200 bp from genomic DNA (Acc.#: XM_113395). Forward primer: 5'-GGT ATG GAT GAG GAG CTG GA, bases from 142 to 161. Reverse primer: 5'-CTT CCA CAG CCC TCG ACT AA, bases from 243 to 224.
This PCR is useful for analysis of DNase digestion as well as cDNA synthesis efficiency.
Picture was kindly provided by Dr. J.-H. Martens, University of Heidelberg. |
||||||||||||||||||||||||||||||||||
| \METHODS\PCR\PCR for PDH | ||||||||||||||||||||||||||||||||||
| © Copyright 1999-2006 Alexei Gratchev. All rights reserved. | ||||||||||||||||||||||||||||||||||