| HOME | METHODS | LINKS | NEWS | POLLS | FORUM | CONTACT | SEARCH |
| Creating a deletion by PCR splicing |
|
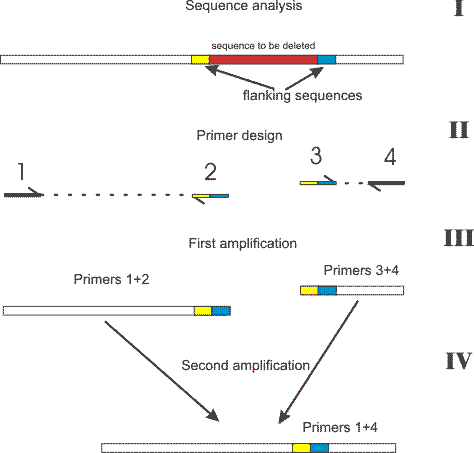
This approach can be used to delete a fragment of any lenth or to introduce point mutations into a DNA seqence. To delete a desired fragment from existing DNA fragment all you need is a pair of primers flanking the region where the deletion will be made (primers 1 and 4), 2 compltementary primers comprising a region of -15 bp to +15 bp related to the junction point (primers 2 and 3) and a high fidelity polymerase (a mix of Taq and Pfu for example). The procedure is shown on the picture. It is always helpful to create expected sequence using any sequence editing software first and then choose primers using this "virtual" construct. Following is important for carrying out the experiment:
 |
| \METHODS\Mutagenesis\PCR splicing |
| © Copyright 1999-2006 Alexei Gratchev. All rights reserved. |
